Lawren Smithline
Email:
lawren@math.cornell.edu
Office: 440 Malott
Hall
Phone: 607-255-8262
Fax: 607-255-7149
13 11 03
Here
is a writeup of the method applied below to Mus Musculus zfp111 and
zfp235.
06 10 03
Here
is the (developing) story of comparing Mus Musculus zfp111 and zfp235.
26 03 03
Here
is an example of another derivative of the TKF
model showing a pair of sequences with multiple regions
of good alignment, in different permutations.
08 03 03
Below is
a representation of possible alignments of 3.5 hectobase segments from
two related genomes. The data came from the Whitehead
Institute. The new method is a derivative of the TKF Markov
model (minus most of the reestimation bells and whistles). The method reports
aligments as a distribution, rather than a single best result.
The deepest red is the most probable.
Prob(alignment passes through a particular purple point)
is less than
exp(-50).Prob(alignment passes through a particular red point).
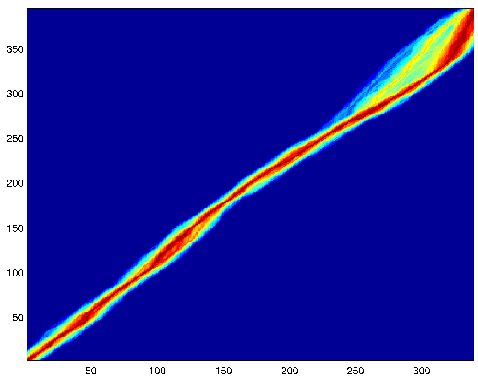
Here's another picture showing the same two sequences aligned with a
related model.

The differences are analogous to cancelling the gap penalty for terminal
gaps. This is expressed by running the TKF model as the sequence extension
process instead of the insertion process along the trailing edges of the
array. This picture shows that this modified TKF process prefers
a terminal gap. The other modification to the this process was the assumption
that the sequences are selections from essentially infinite strings. This
causes a bias away from shortest alignment, or removes a bias towards substitution
events.
lawren@math.cornell.edu
Last modified: Mar 26 11:22:19 EST 2003

