


Next: Alignment of zinc finger
Up: research
Previous: research
- G
-
O. Gotoh,
An improved algorithm for matching biological sequences.
Journal of Molecular Biology 162 (1982) 705-708.
- GM
-
F. Gouvêa and B. Mazur, On the characteristic series of the U
operator. Ann. Inst. Fourier 43 (1993) 301-312.
- H
-
J. Hein, An algorithm for statistical alignment of
sequences related by a binary tree. Preprint.
- H+
-
Hein, J, C. Wiuf, B. Knudsen, M. B. Møller, and G. Wibling,
``Statistical alignment: computational properties, homology testing and
goodness-of-fit.'' J. Mol. Biol. 302:265-279 (2000).
- Ki
-
L. J. P. Kilford, Slopes of overconvergent modular forms.
Ph. D. thesis. Imperial College of Science, Technology and Medicine,
London (2002).
- LGS
-
C. Lee, C. Grasso, M. Sharlow,
Multiple sequence alignment using partial order graphs.
Bioinformatics 18 (2002) 452-464.
- L+
-
Lunter, G. A., I. Miklós, Y. S. Song, and J. Hein, ``An efficient
algorithm for statistical multiple
alignment on arbitrary phylogenetic trees.'' Preprint (April 2003).
- NHH
-
C. Notredame, D. G. Higgins, and J. Heringa,
T-coffee: a novel method for fast and accurate multiple sequence
alignment. Journal of Molecular Biology 302 (2000) 205-217.
- SN
-
N. Saitou and M. Nei,
The neighbor-joining method: a new method for
reconstructing phylogenetic trees.
Molecular Biology and Evolution 4 (1987) 406-425.
- SW
-
T. F. Smith and M. S. Waterman,
Identification of common molecular subsequences.
Journal of Molecular Biology 147 (1981) 195-197.
- Sm
-
L. M. Smithline, Exploring slopes of
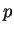 -adic modular forms.
Ph. D. thesis. University of California, Berkeley (2000).
-adic modular forms.
Ph. D. thesis. University of California, Berkeley (2000).
- Sm2
-
--, Compact operators with rational generation. Proceedings of
the Canadian Number Theory Association VII, Montreal (to appear).
- Sm3
-
--, Bounding slopes of
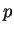 -adic modular forms. Submitted.
-adic modular forms. Submitted.
- Sm4
-
--, Computing lowest slopes of
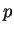 -adic modular forms. Submitted.
-adic modular forms. Submitted.
- Sm5
-
--. Probabilistic pairwise sequence alignment. Preprint.
- TKF
-
J. L. Thorne, H. Kishino, and J. Felsenstein,
An evolutionary model for maximum likelihood alignment
of DNA sequences. Journal of Molecular Evolution 33 (1991) 114-124.
Lawren Smithline
2003-11-29