|
We compare two zinc finger proteins from Mus musculus. We use the mRNA sequences for Zfp111 and Zfp235 [SHGBS]. These two proteins have a similar amino acid sequences: one KRAB domain, followed by a spacer region, followed by a series of zinc finger domains in tandem. The members of this protein family each have from five to nineteen of the 28 amino acid zinc finger domains. Most of these amino acids are required for zinc binding, and are highly conserved between duplications and between genes. Shannon et al. show statistically that over all nucleotide positions in the zinc finger domains, substitution events tend to be synonymous. They observe a range of selection behaviors at positions believed to be noncritical to the zinc binding function.

|
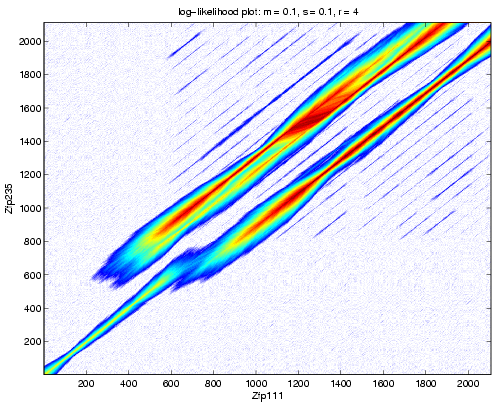
Figure 4 shows the ![]() array computed
with the Zfp111 and Zfp235 sequences. The
array computed
with the Zfp111 and Zfp235 sequences. The ![]() array shows high sequence
identity at the beginning of the sequence, followed by
a mismatch with a net insertion in the Zfp235 sequence.
array shows high sequence
identity at the beginning of the sequence, followed by
a mismatch with a net insertion in the Zfp235 sequence.
|
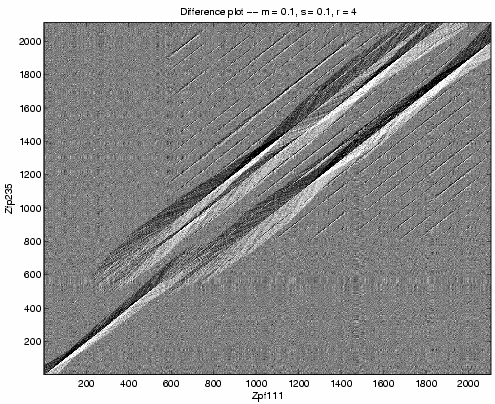
Figure 5 shows the ![]() plot for
the same sequences and parameters. The
plot for
the same sequences and parameters. The ![]() plot
provides more detail on the mismatch discovered by the
plot
provides more detail on the mismatch discovered by the ![]() array plot. We see insertion of the second Zfp235 zinc finger
relative to Zfp111 and insertion of either the
eighth or ninth Zfp111 zinc finger relative to Zfp235.
array plot. We see insertion of the second Zfp235 zinc finger
relative to Zfp111 and insertion of either the
eighth or ninth Zfp111 zinc finger relative to Zfp235.
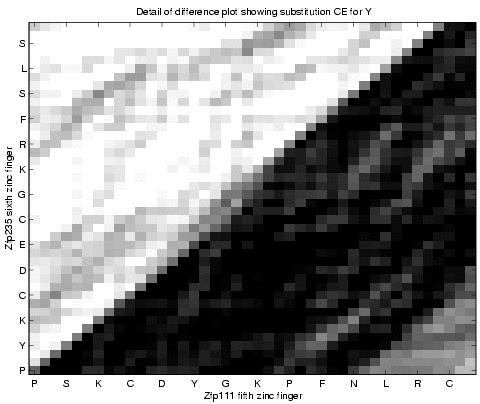
Figure 6 shows a detail of the same ![]() array. We can locate a single amino acid deletion in Zfp111
relative to Zfp235.
array. We can locate a single amino acid deletion in Zfp111
relative to Zfp235.
|
|
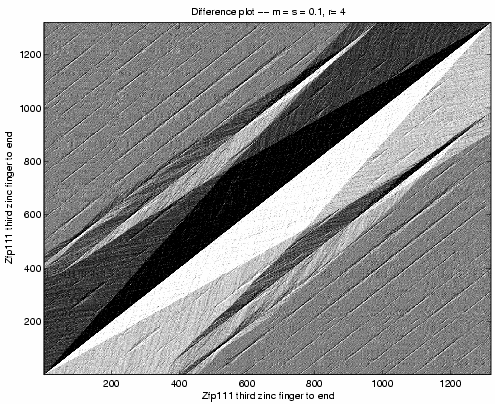
Zinc fingers 10 through 13 of Zfp235 match zinc
fingers 10 through 13 and also zinc fingers 14 through 17
of Zfp111. Figure 7, showing the ![]() array for Zfp111 against itself, provides strong evidence
of the internal duplication in Zfp111.
array for Zfp111 against itself, provides strong evidence
of the internal duplication in Zfp111.